Welcome to Straight Talk about AI in Healthcare. Each post explains recent AI research in non-technical terms and highlights lessons for healthcare organizations, focusing on practical insights to help you understand what you can do today and what should be on your radar for tomorrow.
The discovery of a novel antibiotic using AI was recently announced in various media outlets. This work, published last month by researchers from MIT, demonstrates the potential of AI to drastically accelerate the discovery of new drugs.
According to the CDC, close to 3 million antibiotic-resistant infections occur in the US each year, resulting in more than 35,000 deaths. At the same time, discovering new antibiotics is becoming increasingly difficult. One reason is that most new antibiotics discovered turn out to be very similar to existing antibiotics, so they are not effective against antibiotic-resistant bacteria.
The goal of the MIT study was to identify antibiotic molecules that are substantially different from known antibiotics using deep learning, a family of methods inspired by the way neurons in the brain work. Deep learning methods have been particularly successful in text and image analysis, thanks in part to specific techniques tailored to these areas. By contrast, few of the deep learning methods developed so far are specifically tailored to biological applications. This may be one reason why, as regular readers already know, most applications of deep learning to biological data generally haven’t found wide success.
But, for data about chemical properties of molecules, tailored deep learning methods may be beginning to mature. Specifically, a method called “graph neural network”, which encodes information about the graphical representation of molecules, outperforms both generic deep learning methods and more traditional methods to analyze chemical data.
In this study, the researchers used a library of 2560 molecules, including known antibiotics, other drugs, and natural products. They tested, in the lab, which of these molecules inhibit the growth of E. coli, meaning that the bacteria can’t survive in their presence. They used this data to build a graph neural network that predicts growth inhibition for any given molecule.
Then, the researchers applied this model to a library of over 6000 molecules that are currently under clinical investigation for treating human disease. They examined which molecules were both predicted to inhibit E. coli growth and were dissimilar from known antibiotics.
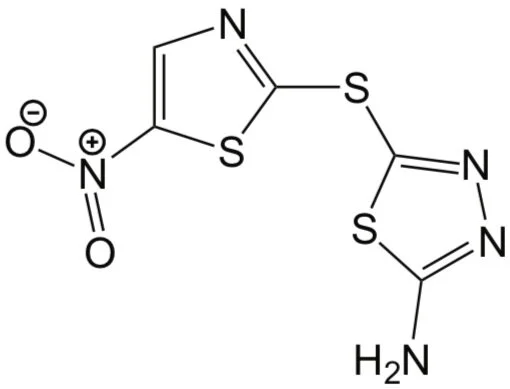
They named the most promising one halicin (see figure), and further lab experiments showed that it is also highly potent against various antibiotic-resistant bacteria. In addition, applying the model to a much larger library of over 100 million molecules, the researchers identified 23 new antibiotic candidates, and validated the activity of the two most promising ones against drug-resistant bacteria.
As with every drug candidate, halicin will have to go through rigorous clinical trials before being approved. Nevertheless, this work shows how AI can dramatically accelerate the discovery of novel drug candidates, especially ones whose structure is highly dissimilar from known antibiotics.
What can you learn from this? Deep learning methods are maturing more quickly for chemical data than genetic data. Data science teams at biopharma companies can expect strong results from applying and scaling these methods in production and not just in academic research settings. However, it is important to recognize that these methods are not fully mature yet: there is no out of the box deep learning technique that “understands chemistry”. AI projects still depend heavily on timely results from the lab, so the most critical ingredient for their success remains tight collaboration between bench scientists and the computational teams.
Not on the mailing list? Subscribe to the newsletter!